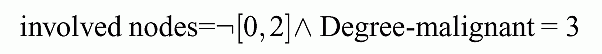
The Mushroom Guide clearly states that there is no simple rule for determining the edibility of these mushrooms; no rule like “leaflets three, let it be“ for Poisonous Oak and Ivy.

8124 cases, 22 symbolic attributes, up to 12 values each, equivalent to 118 logical features.
2480 missing values for attribute 11
51.8% represent edible, the rest non-edible mushrooms.
Safe rule for edible mushrooms:
| odor = (almond.or.anise.or.none) Ù spore-print-color = Ø green | 48 errors, 99.41% correct |
| This is why animals have such a good sense of smell! | |
| Other odors: creosote, fishy, foul, musty, pungent or spicy | |
Rules for poisonous mushrooms - 6 attributes only | |
| R1) odor = Ø (almond Ú anise Ú none); | 120 errors, 98.52% |
| R2) spore-print-color = green | 48 errors, 99.41% correct |
| R3) odor = none Ù
stalk-surface-below-ring = scaly Ù stalk-color-above-ring = Ø brown | 8 errors, 99.90% |
| R4) habitat = leaves Ùcap-color = white | no errors! |
R1 + R2 are quite stable, found even with 10% of data;
R3 and R4 may be replaced by other rules:
R'3): gill-size = narrow Ù stalk-surface-above-ring = (silky Ú scaly)
R'4): gill-size = narrow Ù population = clustered
Only 5 attributes used ! So far the simplest rules.
100% also in crossvalidation tests - structure of this data is completely understandable.
What chemical receptors in the nose realize such discrimination? What does it tell us about evlolution?
Other methods:
| Method | Acc. % |
Rules/Cond Features | Type | Reference |
| RULENEG | 91.0 | 300/8087/? | C | Hayward et.al. |
| HILLARY | 95.0 | ? | C | ML induction, Iba et.al. |
| STAGGER | 95.0 | ? | C | ML induction, Schlimmer |
| REAL | 98.0 | 155/6603/? | C | Craven+Shavlik |
| RULEX | 98.5 | 1/3/? | C | Andrews+Geva |
| DEDEC | 99.8 | 26/26/? | C | Tickle et.al. |
| C4.5 | 99.8 | 3/3/? | C | Quinlan |
| Successive Regularization | 99.4 | 1/4/2 | C | Ishikava |
| 99.9 | 2/22/4 | C | Ishikava | |
| 100 | 3/24/6 | C | Ishikava | |
| TREX | 100 | 3/13/? | F | Geva |
| C-MLP2LN, SSV | 98.5 | 1/3/1 | C | Duch et.al. |
| 99.4 | 2/4/2 | C | Duch et.al. | |
| 99.9 | 3/7/4 | C | Duch et.al. | |
| C-MLP2LN | 100 | 4/9/6 | C | Duch et.al. |
| SSV | 100 | 4/9/5 | C | Duch et.al. |
Artificial small problems designed to test machine learning algorithms (Thurn et.al. 1991).
6 features, 432 possible combinations.
Problem Monk 1:
head shape = body shape OR jacket color = red
124 cases randomly selected for training.
Problem Monk 2:
exactly two of the six features have their first values
169 cases randomly selected for training.
Problem Monk 3:
NOT (body shape = octagon OR jacket color = blue) OR (holding = sward AND jacket color = green)
122 cases randomly selected for training, 5% misclassifcations added.
Such artificial data are difficult to handle.
2 neurons must be trained in C-MLP2LN network simultaneously in Monk 1.
4 neurons must be trained in C-MLP2LN network simultaneously in Monk 2.
Initial rules are too general covering cases from a wrong class.
Exceptions to the general rules: neurons with a negative contribution to the output.
Hierarchical rules: first check exceptions, if not true than rules.
Monk-1: 4 rules and 2 exceptions, 14 atomic formulae.
Monk-2: 16 rules and 8 exceptions, 132 atomic formulae.
Monk-3: 3 rules and 4 exceptions, 33 atomic formulae, 100% accuracy.
Fuzzy methods give poor results here.
| Method | Monk-1 | Monk-2 | Monk-3 | Remarks |
| AQ17-DCI | 100 | 100 | 94.2 | Michalski |
| AQ17-HCI | 100 | 93.1 | 100 | Michalski |
| AQ17-GA | 100 | 86.8 | 100 | Michalski |
| Assistant Pro. | 100 | 81.5 | 100 | Monk paper |
| mFOIL | 100 | 69.2 | 100 | Monk paper |
| ID5R | 79.7 | 69.2 | 95.2 | Monk paper |
| IDL | 97.2 | 66.2 | -- | Monk paper |
| ID5R-hat | 90.3 | 65.7 | -- | Monk paper |
| TDIDT | 75.7 | 66.7 | -- | Monk paper |
| ID3 | 98.6 | 67.9 | 94.4 | Monk paper |
| AQR | 95.9 | 79.7 | 87.0 | Monk paper |
| CLASSWEB 0.10 | 71.8 | 64.8 | 80.8 | Monk paper |
| CLASSWEB 0.15 | 65.7 | 61.6 | 85.4 | Monk paper |
| CLASSWEB 0.20 | 63.0 | 57.2 | 75.2 | Monk paper |
| PRISM | 86.3 | 72.7 | 90.3 | Monk paper |
| ECOWEB | 82.7 | 71.3 | 68.0 | Monk paper |
| Neural methods | ||||
| MLP | 100 | 100 | 93.1 | Monk paper |
| MLP+reg. | 100 | 100 | 97.2 | Monk paper |
| Cascade correlation | 100 | 100 | 97.2 | Monk paper |
| FSM, Gaussians | 94.5 | 79.3 | 95.5 | Duch et.al. |
| SSV | 100 | 80.6 | 97.2 | Duch et.al. |
| C-MLP2LN | 100 | 100 | 100 | Duch et.al. |
286 cases, 201 no recurrence cancer events (70.3%), 85 are recurrence (29.7%) events.
9 attributes, symbolic with 2 to 13 values.
Single rule:
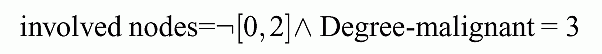
with ELSE condition gives over 77% in crossvalidation;
best systems do not exceed 78% accuracy (insignificant difference).
All knowledge contained in the data is:
IF more than 2 nodes were involved AND cancer is highly malignant THEN there will be recurrence.
C-MLP2LN more accurate rules: 78% overall accuracy
R1: deg_malig=3 & breast=left & node_caps=yes
R2: (deg_malig=3 OR breast=left) & NOT inv_nodes=[0,2] & NOT age=[50,59]
1 % gained - statistically insignificant difference - but much more complex rules.
| Method | | Reference |
| C-MLP2LN | | our |
| CART | | Weiss, Kapouleas |
| PVM | | Weiss, Kapouleas |
| AQ15 | | Michalski et.al |
| Inductive | | Clark, Niblett |
699 cases, 458 benign (65.5%), 241 (34.5%) malignant.
9 features (properties of cells), integers 1-10, one attribute missing in 16 cases.
Simplest rules from C-MLP2LN, large regularization:
IF f2 ł 7 Ú f7 ł 6 THEN malignant (95.6%)
Overall accuracy (including ELSE condition) is 94.9%.
f2 - uniformity of cell size; f7 - bland chromatin
Hierarchical sets of rules with increasing accuracy may be build
More accurate set of rules:
| R1: f2<6 Ù f4<3 Ù f8<8 | (99.8)% |
| R2: f2<9 Ù f5<4 Ù f7<2 Ù f8<5 | (100)% |
| R3: f2<10 Ù f4<4 Ù f5<4 Ù f7<3 | (100)% |
| R4: f2<7 Ù f4<9 Ù f5<3 Ù f7Î [4,9] Ù f8<4 | (100)% |
| R5: f2Î [3,4]Ù f4<9 Ù f5<10 Ù f7<6 Ù f8<8 | (99.8)% |
R1 and R5 misclassify the same 1 benign vector.
ELSE condition makes 6 errors, overall reclassification accuracy 99.00%
In all cases features f3 and f6 (uniformity of cell shape and bare nuclei) are not important, f2 (clump thickness) and f7 (bare nuclei) being the most important.
100% reliable set of rules rejects 51 cases (7.3%).
Results from the 10-fold (stratified) crossvalidation - accuracy of rules is hard to compare without the test set
| Method | % accuracy |
| IncNet | 97.1 |
| 3-NN, Manhattan | 97.1± 0.1 |
| Fisher LDA | 96.8 |
| MLP+backpropagation | 96.7 |
| LVQ (vector quantization) | 96.6 |
| Bayes (pairwise dependent) | 96.6 |
| FSM, 12 fuzzy Gaussian rules | 96.5 |
| Naive Bayes | 96.4 |
| SSV, 3 crisp rules | 96.3±0.2 |
| DB-CART | 96.2 |
| Linear Discriminant Analysis | 96.0 |
| RBF | 95.9 |
| CART (decision tree) | 94.2 |
| LFC, ASI, ASR (decision trees) | 94.4-95.6 |
| Quadratic Discriminant Analysis | 34.5 |
Reclassifcation results are only about 1% better than 10xCV
| Method | Accuracy | Rules/type |
| C-MLP2LN | 99.0 | 5 crisp |
| C-MLP2LN | 97.7 | 4 crisp |
| SSV | 97.4 | 3 crisp |
| NEFCLASS | 96.5 | 4 fuzzy |
| C-MLP2LN | 94.9 | 2 crisp |
| NEFCLASS | 92.7 | 3 fuzzy |
Data from Machine Learning Database repository, UCI
3 classes: hypothyroid, hiperthyroid, normal;
# training vectors 3772 = 93+191+3488
# test vectors 3428 = 73+177+3178
21 attributes (medical tests), 6 continuos
Optimized rules: 4 errors on the training set (99.89%), 22 errors on the test set (99.36%)
| primary hypothyroid: | TSH>30.48 & FTI <64.27 | 97.06% |
| primary hypothyroid: | TSH=[6.02,29.53] & FTI <64.27 & T3< 23.22 | 100% |
| compensated: | TSH > 6.02 & FTI=[64.27,186.71] & TT4=[50, 150.5) &
On_Tyroxin=no & surgery=no | 98.96% |
| no hypothyroid: | ELSE | 100% |
4 continuos attributes used and 2 binary.
| Method | Reference | ||
| C-MLP2LN rules + ASA | | | our group |
| CART | | | Weiss |
| PVM | | | Weiss |
| IncNet | | |
our group |
| MLP init+ a,b opt. | | | our group |
| C-MLP2LN rules | | | our group |
| Cascade correlation | | | Schiffmann |
| BP + local adapt. rates | | | Schiffmann |
| BP+genetic opt. | | | Schiffmann |
| Quickprop | | | Schiffmann |
| RPROP | | | Schiffmann |
| 3-NN, Euclides, 3 features used | | | our group |
| 1-NN, Euclides, 3 features used | | | our group |
| Best backpropagation | | | Schiffmann |
| 1-NN, Euclides, 8 features used | | | our group |
| Bayesian classif. | | | Weiss |
| BP+conjugate gradient | | | Schiffmann |
| 1-NN Manhattan, std data | | our group | |
| default: 250 test errors | | ||
| 1-NN Manhattan, raw data | | our group |
Why logical rules are most accurate here?
Probably doctors assigned patients to crisp classes: hypo, hiper. normal on basis of sharp decisions.
MLP is not able to describe sharp rectangular decision borders unless very large weights or large slopes are used.
Training set 43500, test set 14500, 9 attributes, 7 classes
Approximately 80% of the data belongs to class 1, only 6 vectors in class 6.
Rules from FSM after optimization: 15 rules, train 99.89%, test 99.81% accuracy.
32 rules obtained from SSV give 100% train, 99.99% test accuracy (1 error).
| Method | % training | % test | Reference |
| SSV, 32 rules | 100 | 99.99 | our result, 1 test error |
| NewID decision tree | 100 | 99.99 | Statlog |
| Baytree decision tree | 100 | 99.98 | Statlog |
| CN2 decision tree | 100 | 99.97 | Statlog |
| FSM, 17 rules | 99.98 | 99.97 | our group; 1 test error and 3 unclassfied |
| CART | 99.96 | 99.92 | Statlog |
| C4.5 | 99.96 | 99.90 | Statlog |
| FSM, 15 rules | 99.89 | 99.81 | our group |
| MLP | 95.50 | 99.57 | Statlog |
| k-NN | 99.61 | 99.56 | Statlog |
| RBF | 98.40 | 98.60 | Statlog |
| Logistic DA | 96.06 | 96.17 | Statlog |
| LDA | 95.02 | 95.17 | Statlog |
| Naive Bayes | 95.40 | 95.50 | Statlog |
| Default | 78.41 | 79.16 |
FSM: 17 crisp rules make 3 errors on training (99.99%), 8 vectors are unclassified, no errors on the test, 9 vectors unclassified (99.94%).
Gaussian fuzzification (0.05%): 3 errors + 5 unclassified on training, 3 unclassified and 1 error (with p of correct class close to 50%) on test.
NewID never was the best in StatLog project, os this is probably good luck.
More examples of logical rules discovered are on our rule-extraction WWW page and SSV results page